Large language models (LLMs) have achieved unprecedented performance by leveraging vast pretraining corpora—the "fossil fuel" of modern AI—as predicted by scaling laws. However, the diminishing supply of high-quality, human-annotated data, especially in specialized domains, demands a shift toward synthetic data as a new energy source for further advancements. In this paper, we propose a novel Structural Entropy-guided Knowledge Navigator (SENATOR) framework that addresses the intrinsic knowledge deficiencies of LLMs. Our approach employs the Structure Entropy (SE) metric to quantify uncertainty along knowledge graph paths and leverages Monte Carlo Tree Search (MCTS) to selectively explore regions where the model lacks domain-specific knowledge. Guided by these insights, the framework generates targeted synthetic data for supervised fine-tuning, enabling continuous self-improvement. Experimental results on medical benchmarks demonstrate that our SENATOR agent effectively supplements the pretraining corpus by injecting missing domain-specific information, leading to significant performance gains in models such as Llama-3 and Qwen2. Our findings highlight the potential of synthetic data as the “new energy” for LLMs, paving the way for more efficient and scalable strategies to sustain and enhance model performance.
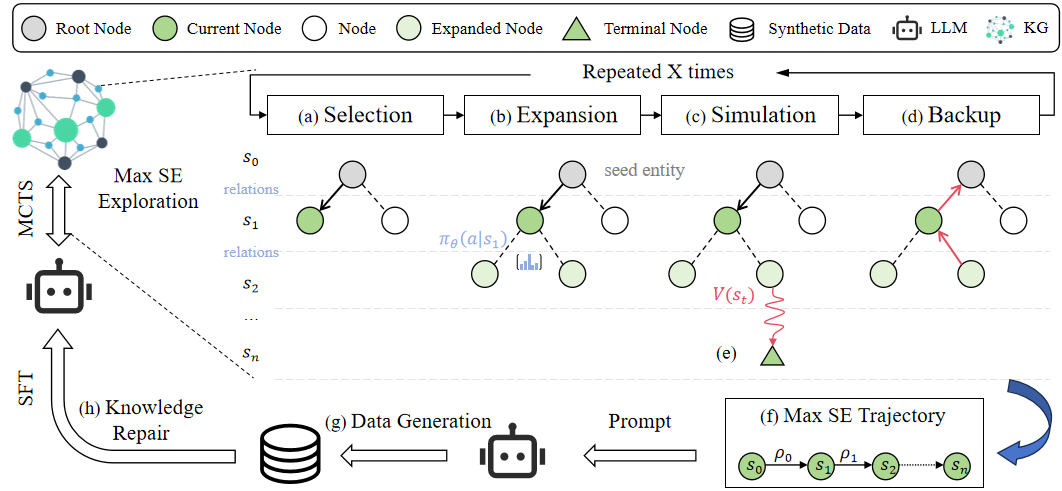
Figure 1: The overall framework of SENATORgit clone https://github.com/weiyifan1023/senator.git
cd senator
conda create -n senator python=3.10.9
conda activate senator
pip install -r requirements.txt
The seed entities of SPOKE KG are derived from the Project KG_RAG
Download the Instruction Tuning dataset from the Paper PMC-LLaMA.
Place the entire ./data/benchmark_data folder under the root folder.
Preprocess your datasets to SFT format by running:
cd llm_rlhf/step1_supervised_finetuning/train_scripts
python preprocessing.pyHere, we initialize subgraph for MCTS, and exploration maximum entropy path.
(Customize the search depth)
python -m prompt_based_generation/MedLLMs/gen_synthetic_data.pyWe take Qwen2-7B as an example, replace some paths in run_qwen.sh, and then execute:
cd llm_rlhf/step1_supervised_finetuning
bash train_scripts/qwen2/run_qwen.shTo eval MedQA, MedMCQA and PubMedQA datasets, you can run:
cd prompt_based_generation/MedLLMs
python eval_medical_qa.pyIf you find our paper inspiring and have utilized it in your work, please cite our paper.
@article{wei2025structural,
title={Structural Entropy Guided Agent for Detecting and Repairing Knowledge Deficiencies in LLMs},
author={Wei, Yifan and Yu, Xiaoyan and Pan, Tengfei and Li, Angsheng and Du, Li},
journal={arXiv preprint arXiv:2505.07184},
year={2025}
}
Thanks to the authors of KG-RAG and DAMe for releasing their code for retrieving the SPOKE KG and evaluating SE on the graph. Much of this codebase has been adapted from their codes.