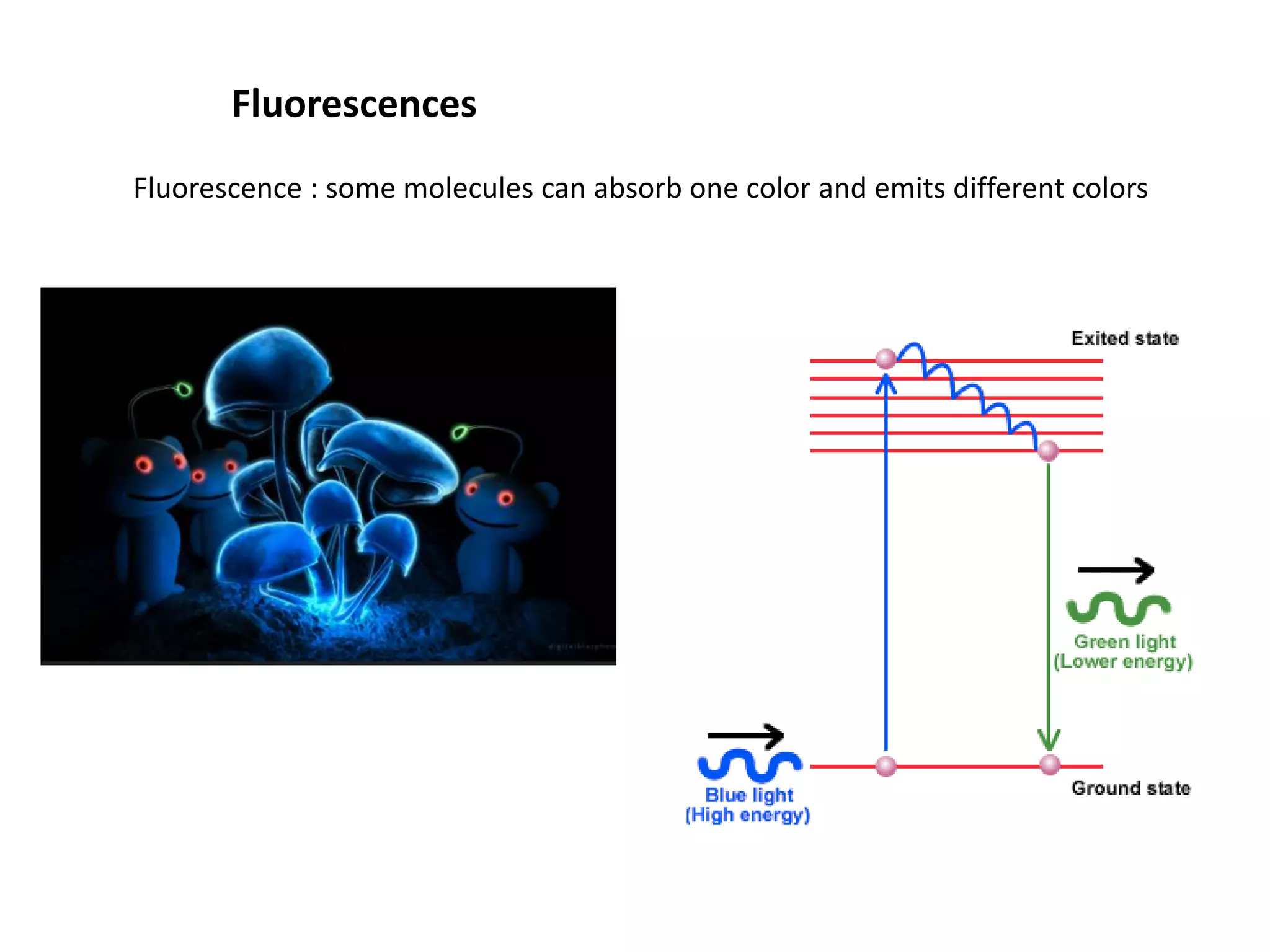
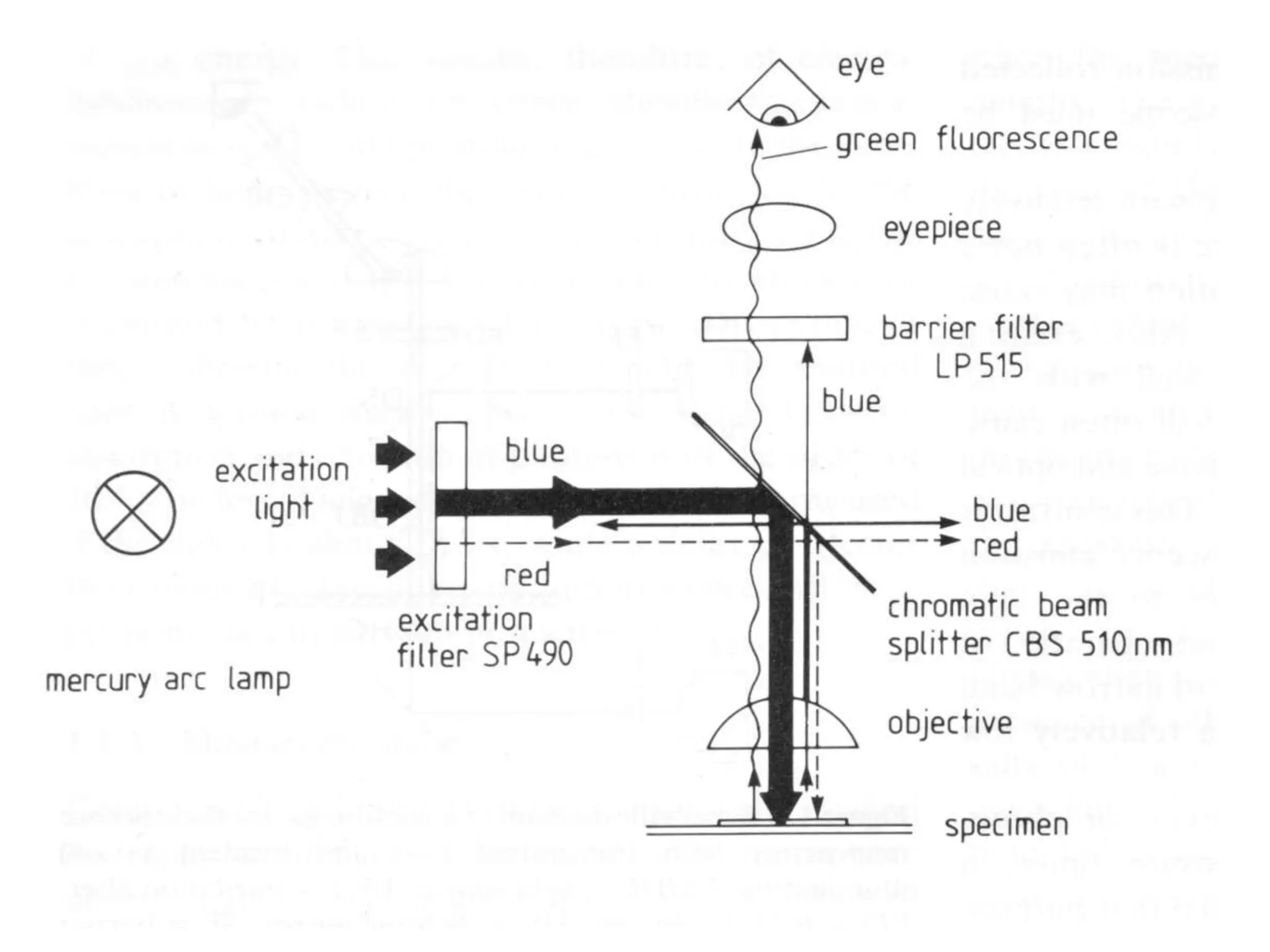
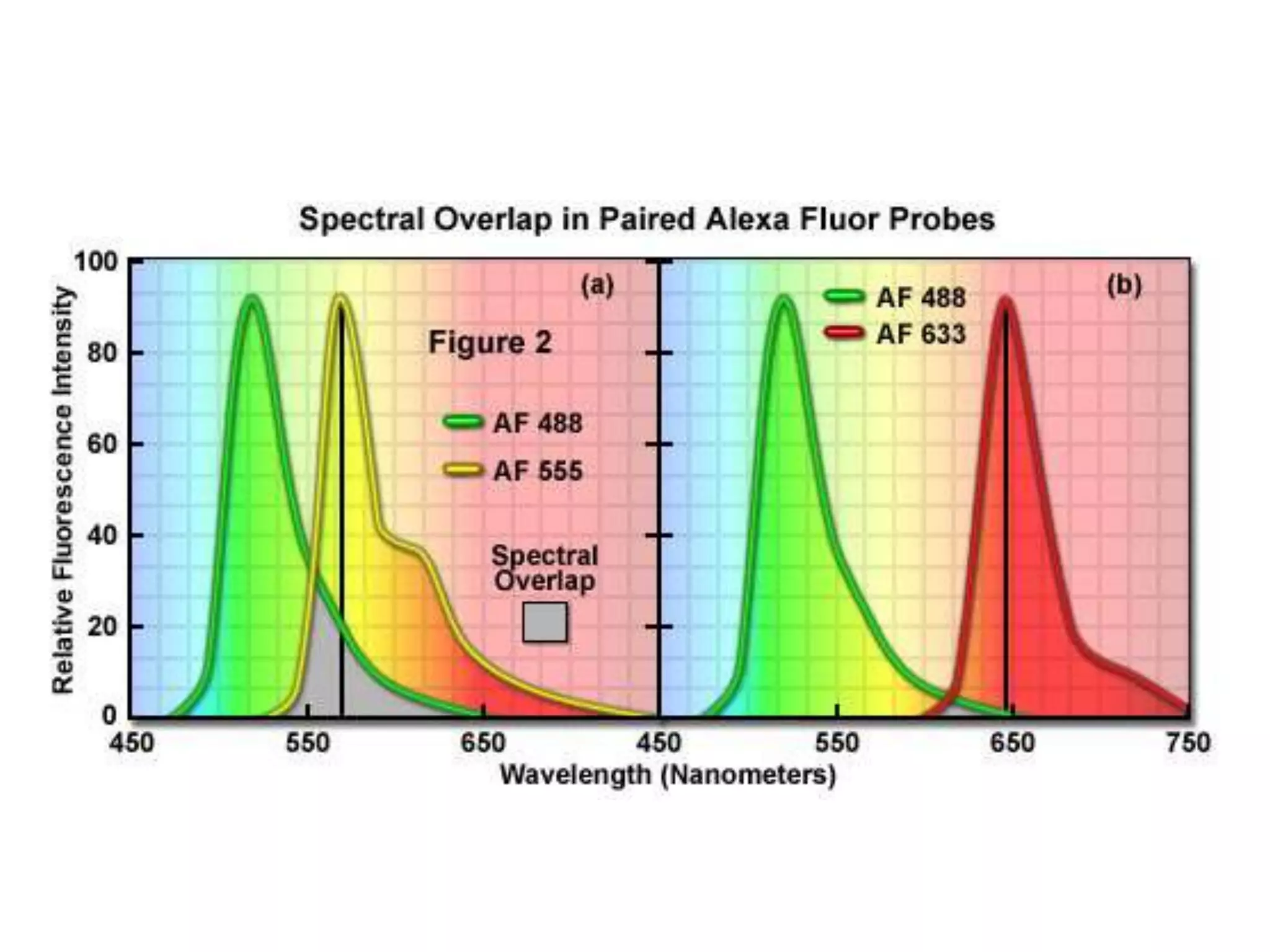
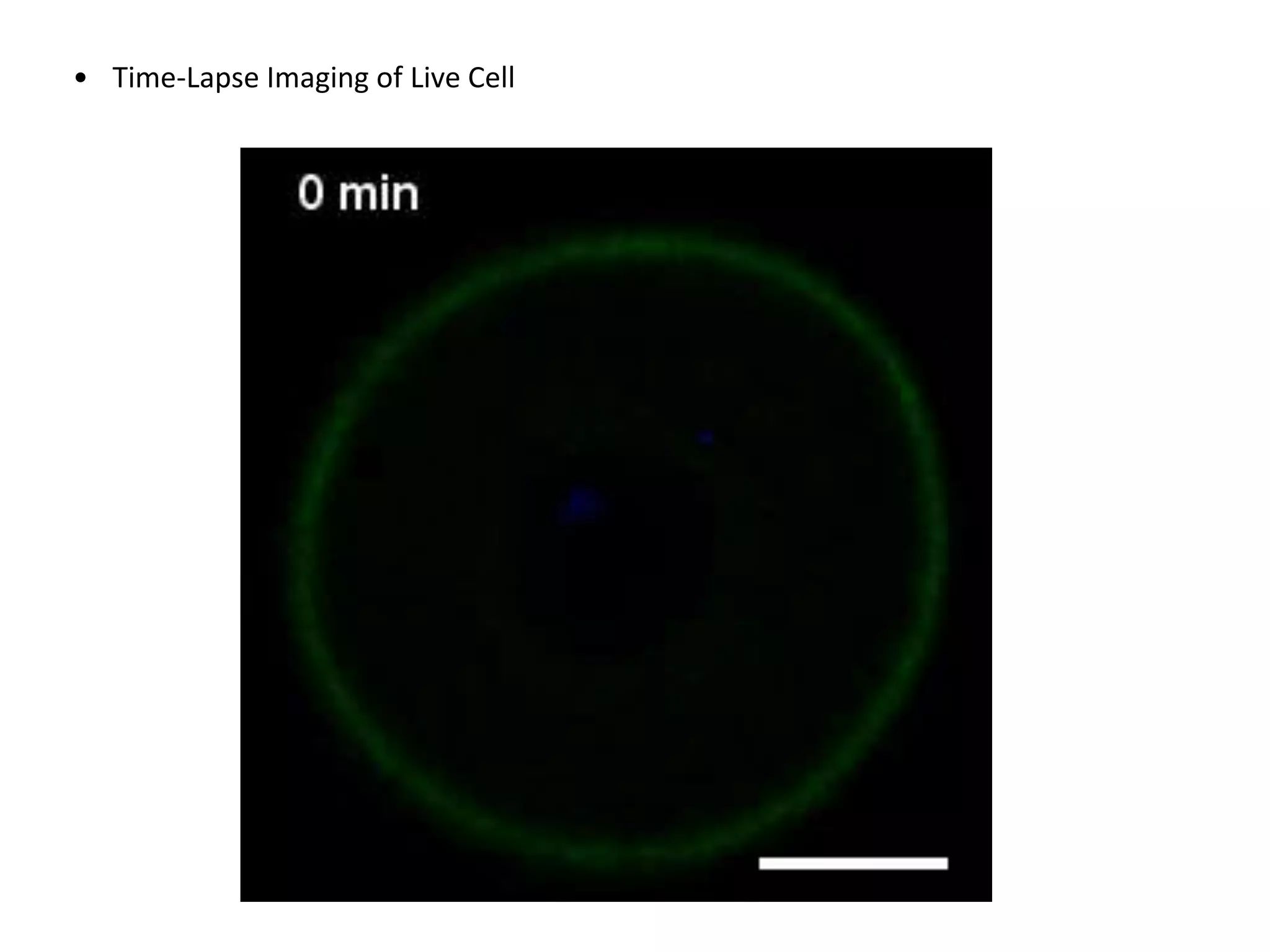
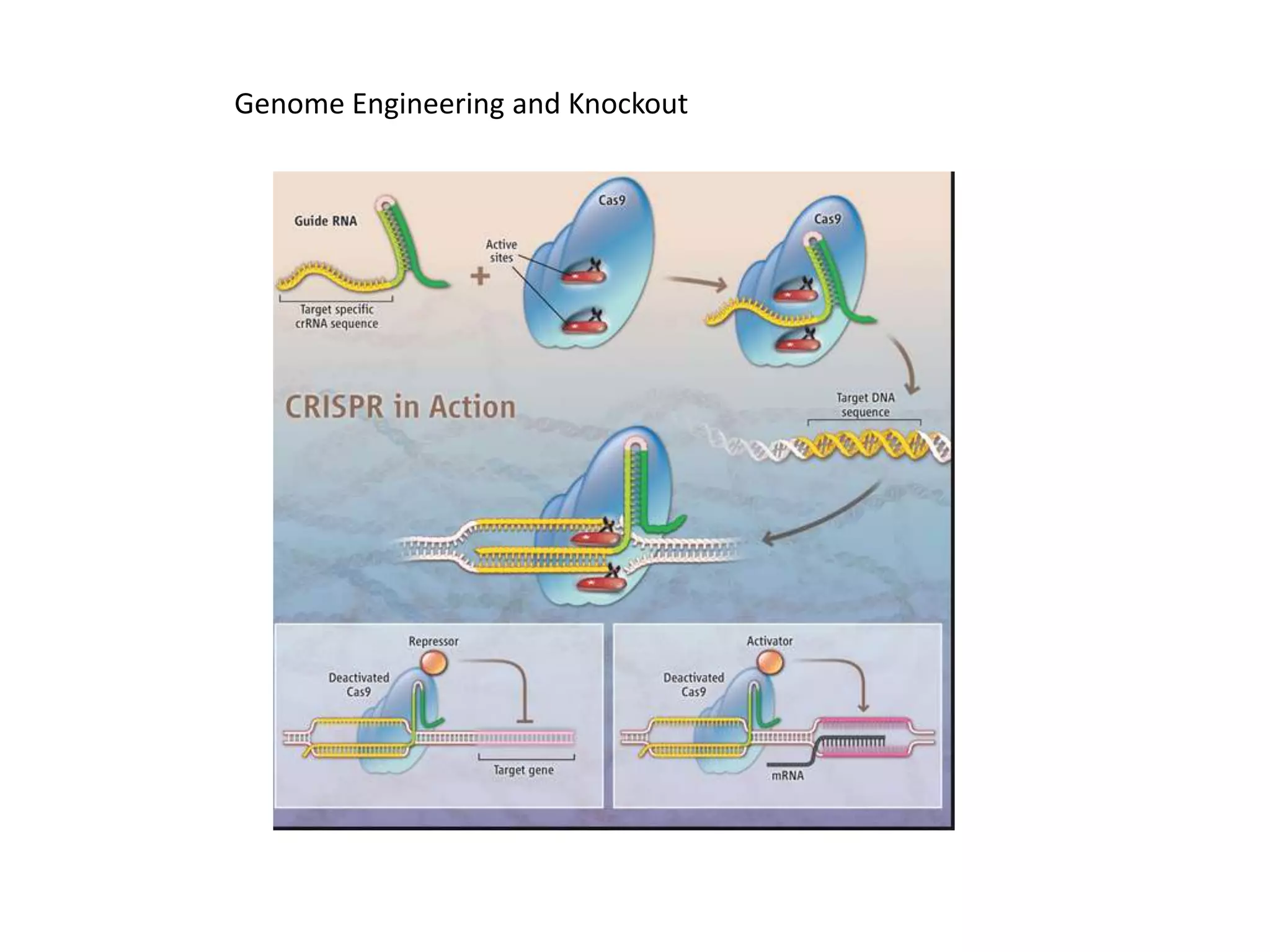
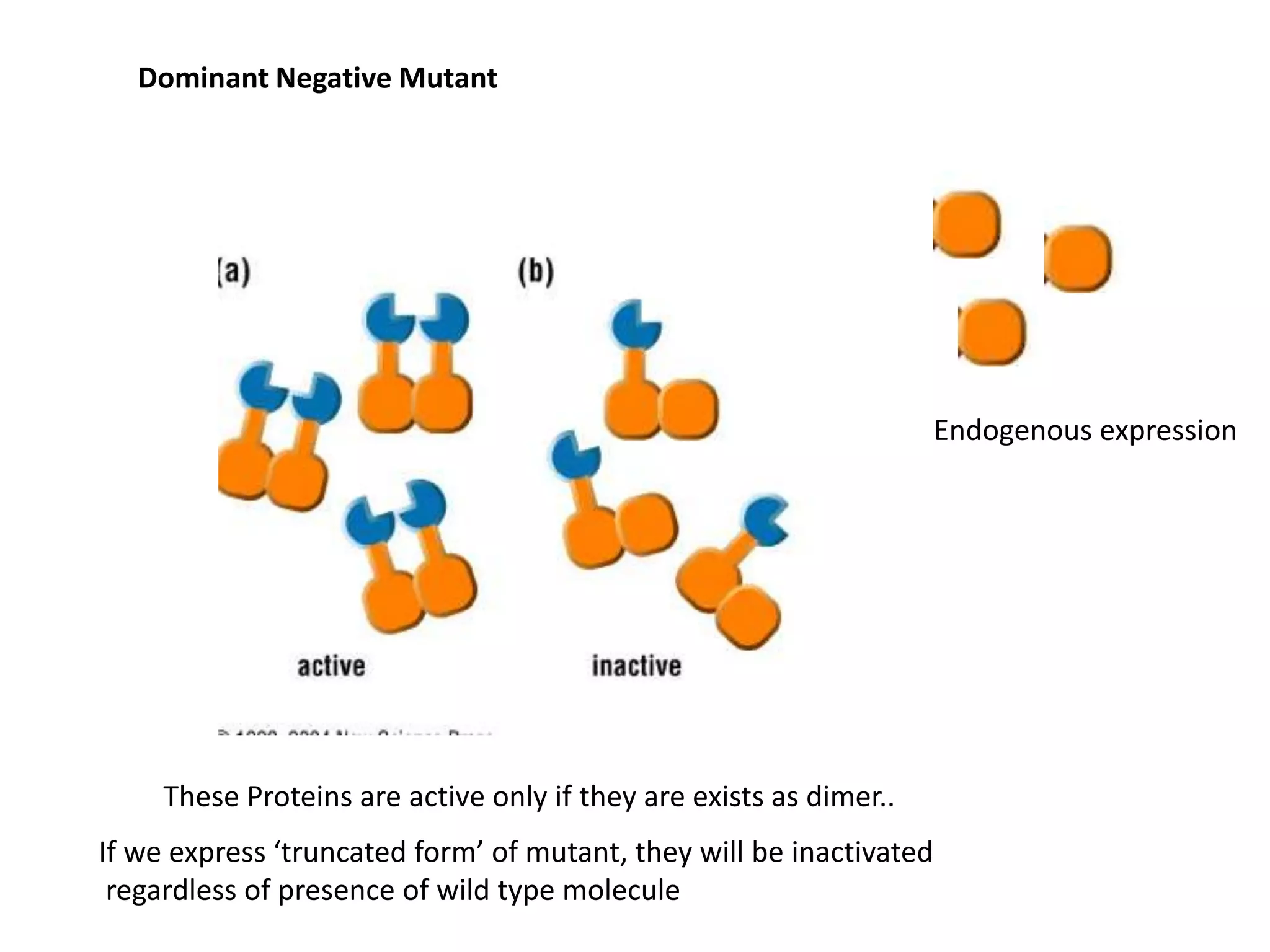
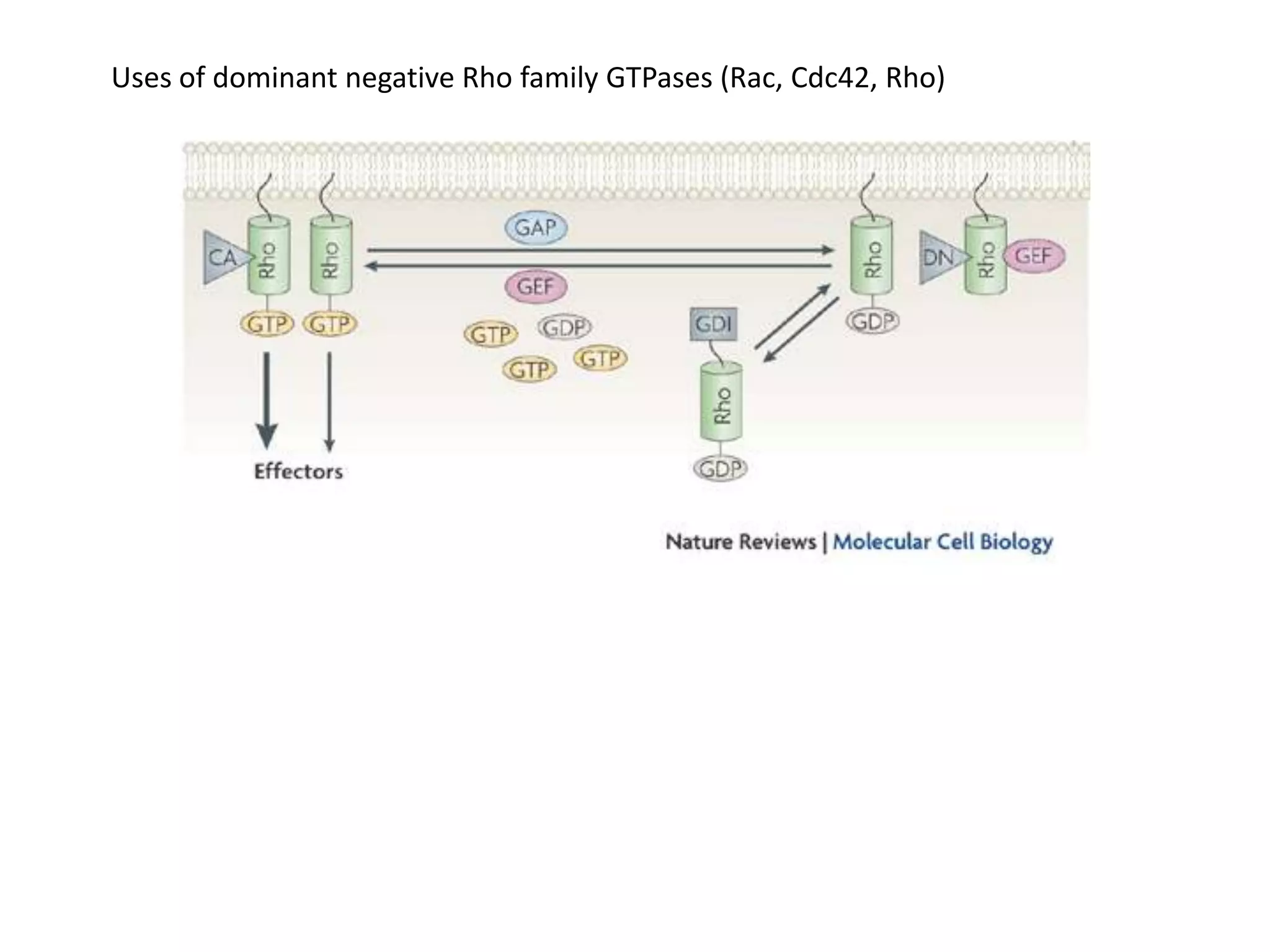
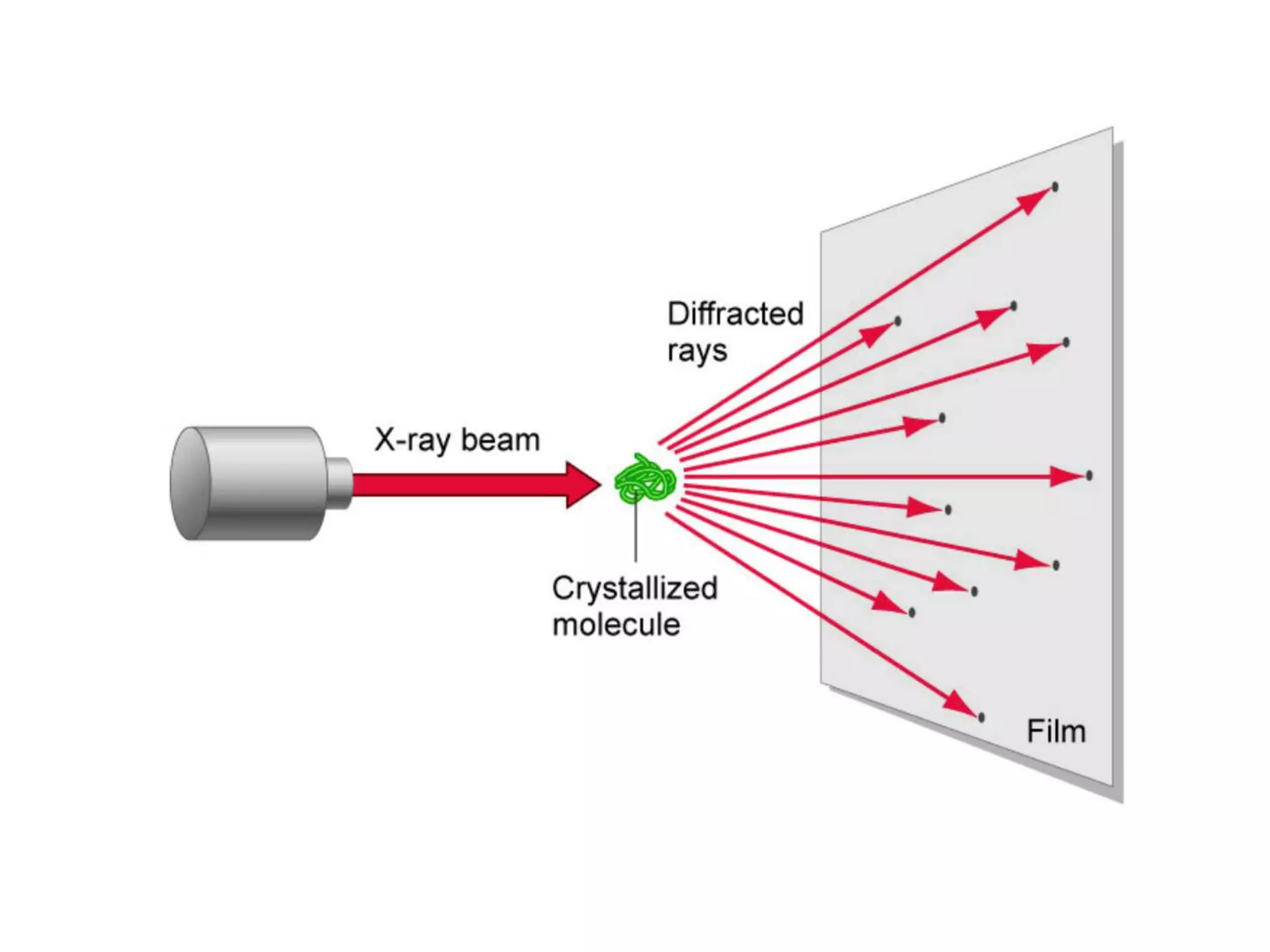
This document provides an overview of advanced cell biology topics covered in a 2014 semester course, including introduction, research strategies, organelles, membranes, trafficking, signaling, cytoskeleton, and cell cycle. It also discusses methods for studying cells, such as determining component lists through expression profiling, proteomics, localization techniques like immunofluorescence, and perturbation methods like RNAi and CRISPR/Cas9 knockouts. Microscopy techniques like fluorescence, confocal, and live cell imaging are explained. The document concludes with an overview of biochemical characterization and protein structure determination using X-ray crystallography.