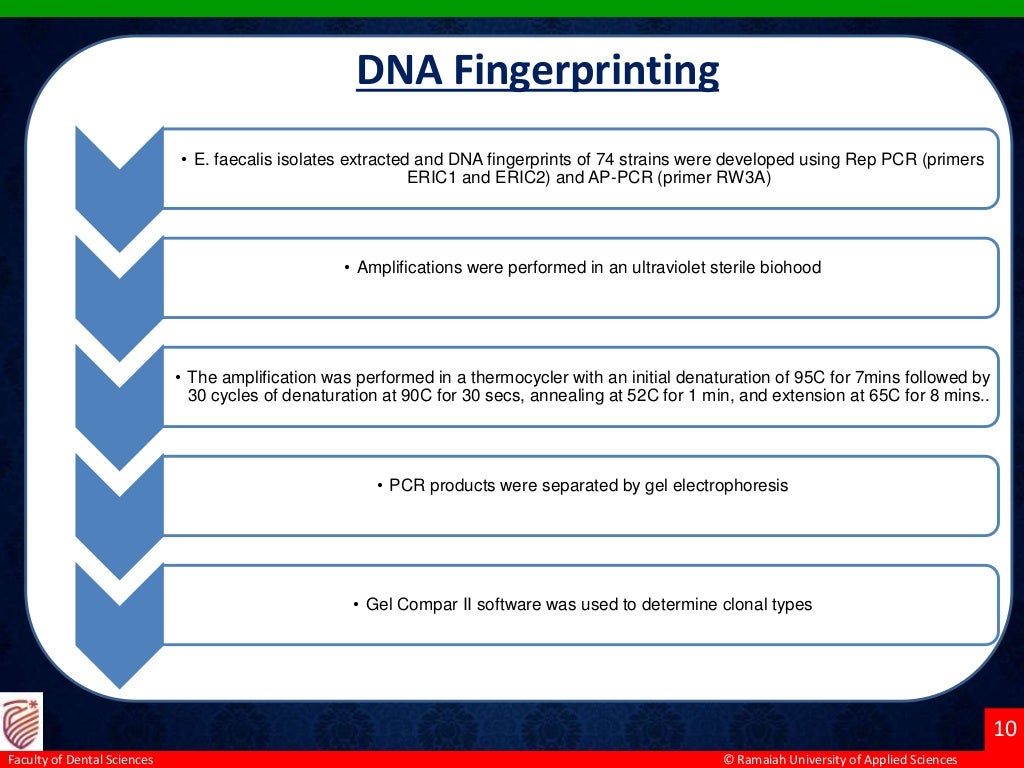
The study aimed to identify and analyze the diversity of Enterococcus faecalis genotypes from multiple oral sites associated with endodontic failure in 20 patients with apical periodontitis. Using in vivo sampling and molecular methods, E. faecalis was isolated from saliva, pulp chambers, and root canals, revealing 7 genotypic groups among 74 strains. The findings suggest a correlation between coronal microleakage and endodontic treatment failure, as similar genotypes were found across different sampled sites.