This PPT inlcudes,
1. Introduction to DNA replication
2. Methods of replication
3. Semiconservative replication
4. Meselson and Stahl experiment
5. Taylor's experiment
6. Replicon and origin of replication
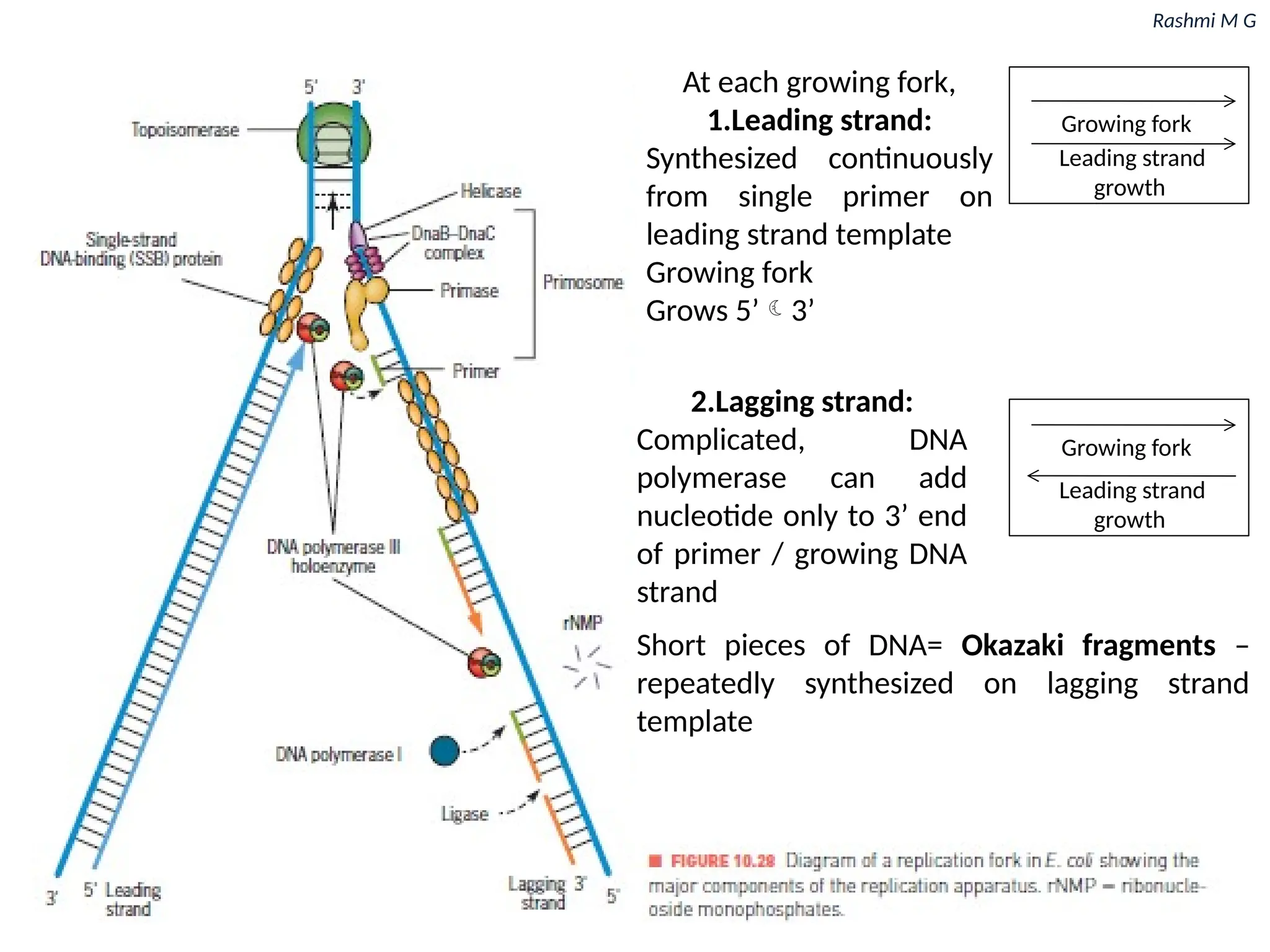
7. DNA replication in prokaryotes
8. Biochemistry of DNA replication
9. Common enzymes involved in DNA replication
10. DNA polymerase
11. Components of DNA Polymerase III
12. Steps involved in prokaryotic DNA replication
13. DNA replication in eukaryotes
14. Unique aspects of eukaryotic DNA replication
15. Eukaryotic DNA Polymerases
16. Proof reading
17. Comparison between DNA replication in Prokaryotes and Eukaryotes